Coronaviruses – a general introduction
March 25, 2020
Who first described them; why they are called coronaviruses; what they are; how they invade cells; how we detect them
Jeffrey K Aronson
Centre for Evidence-Based Medicine, Nuffield Department of Primary Care Health Sciences
University of Oxford
Until recently, most people will never have heard of coronaviruses. But they, and the diseases they cause in humans and animals, have been recognized for over 50 years.
Who first discovered coronaviruses?
Avian infectious bronchitis was first described in newborn chicks in 1931 by Schalk & Hawn (J Am Vet Med Ass 1931; 78: 413–23) and by Bushnell & Brandly in 1933 (Poultry Science 1933; 12: 55-60). These papers were both cited by Beach & Schalm, 1936, who confirmed that the infection was due to a filterable virus and identified two strains, with cross-immunity. The virus was cultivated in 1937 by Fred Beaudette and Charles Hudson, from the New Jersey Agricultural Experiment Station (J Am Vet Med Ass 1937; 90: 51–8 cited by Marks) and later by Cunningham & Stuart in 1947.
In 1951 Gledhill & Andrewes isolated a hepatitis virus from mice, now also known to be a coronavirus.
In 1965, the virologist David Tyrrell, Director of the Medical Research Council’s Common Cold Research Unit at Harnham Down near Salisbury in Wiltshire, and his colleague Mark Bynoe published a paper in the British Medical Journal, in which they described a virus, which they called B814, and identified it as a cause of the common cold. They tried to characterize other viruses, but without much success, and thought that viruses of which they found evidence were rhinoviruses.
On 1 April 1967 Tyrell, this time with his colleague June Almeida, from the Department of Medical Microbiology in London’s St Thomas’s Hospital Medical School, identified three uncharacterized respiratory viruses, of which two had not previously been associated with human diseases. They reported that two of the viruses, 229E and B814, of which they published electron micrographs, were indistinguishable from the particles of avian infectious bronchitis.
Then Almeida and Tyrell, with six other colleagues, reported in Nature in 1968 that there was a group of viruses that caused not only avian bronchitis but also murine hepatitis and upper respiratory tract diseases in humans, as shown in Figure 1, taken from their brief annotation, which was published under the general heading “News and Views” (Almeida JD, Berry DM, Cunningham CH, Hamre D, Hofstad MS, Mallucci L, McIntosh K, Tyrrell DAJ. Virology: Coronaviruses. Nature 1968; 220(5168): 650). This is the first recorded instance of the term “coronaviruses”.
The virus of avian infectious bronchitis is classified as a gammacoronavirus, while most of the coronaviruses that infect humans are betacoronoviruses. The human coronavirus HCoV-229E described by Almeida and Tyrrell is an alphacoronavirus.
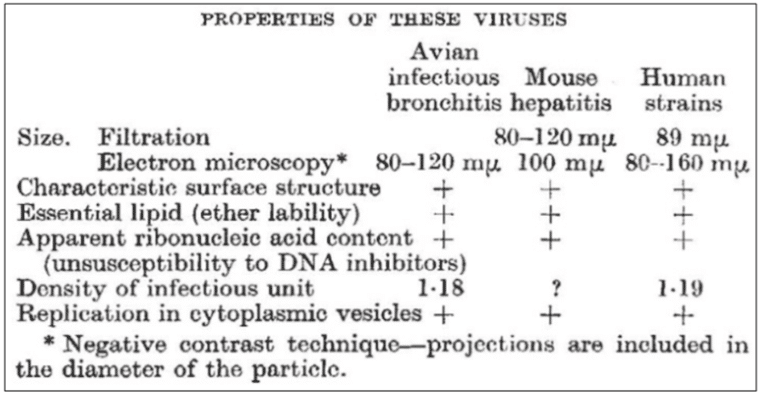
Figure 1. Details of the properties of coronaviruses, first published in Nature (1968; 220(5168): 650); David Tyrrell, at the Common Cold Research Unit, Salisbury, Wiltshire, offered to provide a short bibliography to anyone interested in the data on which the table was based
Why are they called coronaviruses?
As the journal Nature reported in 1968, “these viruses are members of a previously unrecognized group which [the virologists] suggest should be called the coronaviruses, to recall the characteristic appearance by which these viruses are identified in the electron microscope.”
The word “corona” has many different meanings (see Appendix 2). But it was the sun that the virologists had in mind when they chose the name coronaviruses. As they wrote, they compared “the characteristic ‘fringe’ of projections” on the outside of the virus with the solar corona (not, as some have suggested, the points on a crown). Figure 2 illustrates this.
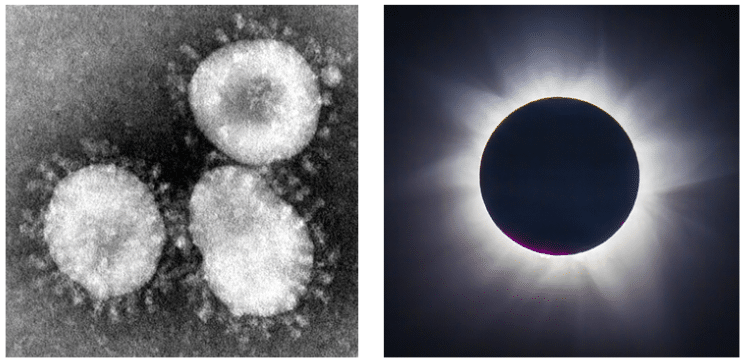
Figure 2. Left: The virions of coronaviruses; Right: The corona of the sun seen during an eclipse
What are coronaviruses and how do they invade cells?
Coronaviruses are single-stranded RNA viruses, about 120 nanometers in diameter. They are susceptible to mutation and recombination and are therefore highly diverse. There are about 40 different varieties (see Appendix 1) and they mainly infect human and non-human mammals and birds. They reside in bats and wild birds, and can spread to other animals and hence to humans. The virus that causes COVID-19 is thought to have originated in bats and then spread to snakes and pangolins and hence to humans, perhaps by contamination of meat from wild animals, as sold in China’s meat markets.
The corona-like appearance of coronaviruses is caused by so-called spike glycoproteins, or peplomers, which are necessary for the viruses to enter host cells. The spike has two subunits; one subunit, S1, binds to a receptor on the surface of the host’s cell; the other subunit, S2, fuses with the cell membrane. The cell membrane receptor for both SARS-CoV-1 and SARS-CoV-2 is a form of angiotensin converting enzyme, ACE-2, different from the enzyme that is inhibited by conventional ACE-1 inhibitors, such as enalapril and ramipril.
Briefly, the S1 subunit of the spike binds to the ACE-2 enzyme on the cell membrane surface. A host transmembrane serine protease, TMPRSS2, then activates the spike and cleaves ACE-2. TMPRSS2 also acts on the S2 subunit, facilitating fusion of the virus to the cell membrane. The virus then enters the cell. Inside the cell the virus is released from endosomes by acidification or the action of an intracellular cysteine protease, cathepsin.
A model and a more detailed description of these events is shown in Figure 3.
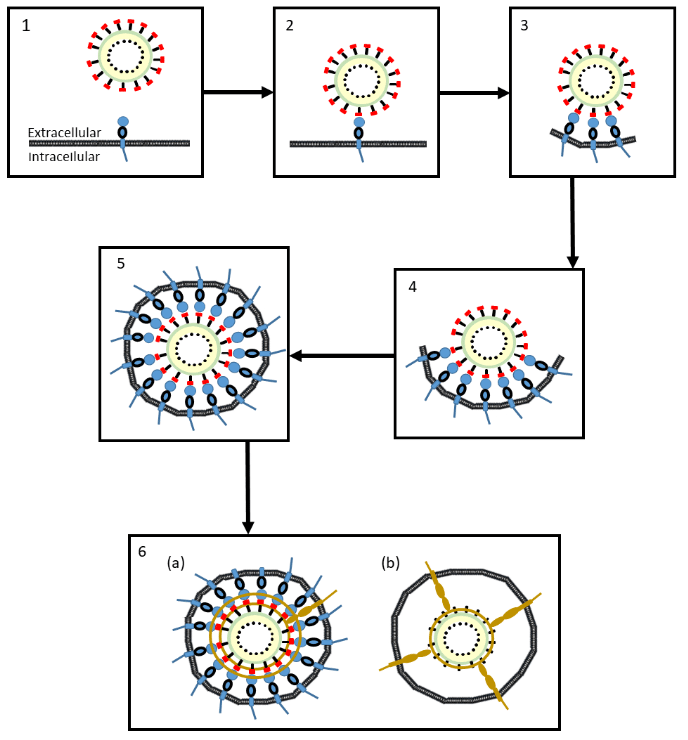
Figure 3. A proposed model of the mechanisms whereby coronavirus SRA-CoV-2 enters cells
- The coronavirus approaches the cell membrane
- An S1 subunit (red) at the distal end of a glycoprotein spike of the virus binds to a membrane-bound molecule of ACE-2 (blue)
- As more S1 subunits of the glycoprotein spikes bind to membrane-bound molecules of ACE-2, the membrane starts to form an envelope around the virus (an endosome)
- The process continues …
- … until the endosome is complete
- The virus can enter the cell in two ways:
(a) A cell membrane-bound serine protease (brown), TMPRSS2, cleaves the virus’s S1 subunits (red) from its S2 subunits (black) and also cleaves the ACE-2 enzymes; the endosome enters the cell (endocytosis), where the virus is released by acidification or the action of another protease, cathepsin
(b) The same serine protease, TMPRSS2, causes irreversible conformational changes in the virus’s S2 subunits, activating them, after which the virus fuses to the cell membrane and can be internalized by the cell
A serine protease inhibitor, camostat mesylate, used in Japan to treat chronic pancreatitis, inhibits the TMPRSS2 and partially blocks the entry of SARS-CoV-2 into bronchial epithelial cells in vitro.
Research interest in coronaviruses
The first coronaviruses found to infect humans were called 229E and OC43, but they caused very mild infections, similar to the common cold. It was not until the outbreaks of SARS (severe acute respiratory syndrome) and then MERS (the Middle Eastern respiratory syndrome or camel flu) that it was appreciated that they could cause serious human infections. Those two infections are thought to have come from bats via civet cats and camels.
This awakening of interest in coronaviruses at different times is reflected in the pattern of publications about them. After the initial description of coronaviruses in 1968 there was a slow increase in the numbers of publications dealing with them, followed by two peaks, after two epidemics: the SARS coronavirus epidemic in 2003–4 and an outbreak of porcine epidemic diarrhoea in North America in 2013 (Figure 4). Identification of the first cases of MERS in Saudi Arabia in 2012, and then elsewhere (e.g. in South Korea in 2015), also caused by a coronavirus, may also have contributed.
I have previously highlighted the fact that the major peaks of interest in the coronaviruses have followed major infections in humans and animals. In my BMJ opinion column on 31 January this year, where some of this article has previously appeared. I wrote that I expected to see another peak in the numbers of publications following the current epidemic. My original graph ended with the 2019 figures. I have now added the latest numbers, from 2020, to the graph, which shows that my prophecy has already been fulfilled. More publications on coronaviruses have been logged in Pubmed in the first 12 weeks of 2020 than in any previous complete year. The difficulty in preventing and treating the infection is matched by the difficulty in keeping up with the published literature.
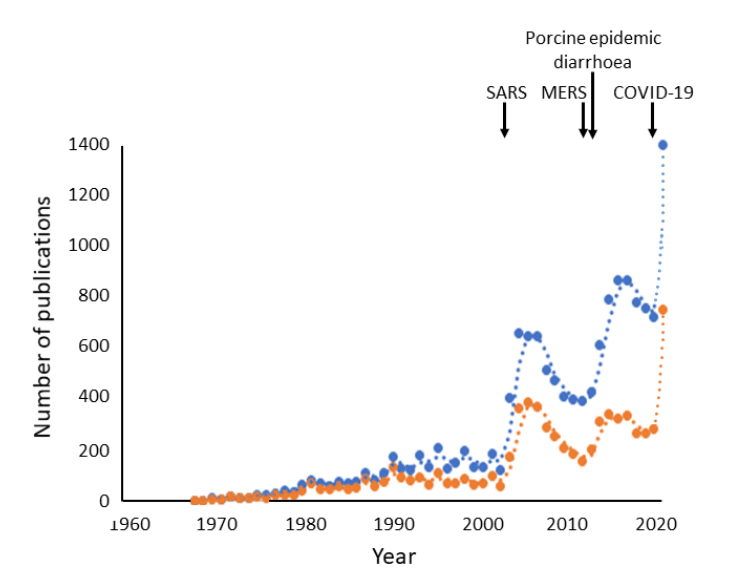
Figure 4. Numbers of publications with “coronavirus/es” as text words (blue) or in titles (orange) (source PubMed Legacy); each point represents one year, but the rightmost points cover only the first 12 weeks of 2020
Testing for coronavirus, SARS-CoV-2
Viral RNA can be detected by polymerase chain reaction (PCR, or quantitative PCR, qPCR, sometimes referred to as “real-time PCR” or RT-PCR, causing confusion with another term, “reverse transcriptase PCR”) (Figure 5). In this test, the virus’s single-stranded RNA is converted to its complementary DNA by reverse transcriptase; specific regions of the DNA, marked by so-called primers, are then amplified. This is done by synthesizing new DNA strands from deoxynucleoside triphosphates using DNA polymerase. Occasional false negatives have been reported.
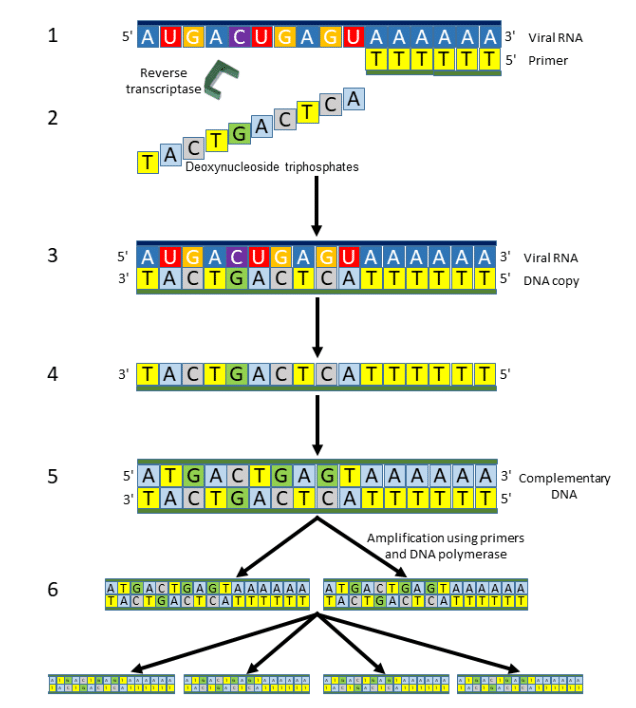
Figure 5. Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
- A primer is attached to the 3 prime end of a single strand of viral RNA
- Deoxynucleoside triphosphates are added stepwise …
- … creating a DNA copy of the viral RNA
- The single strand of DNA is separated …
- … and double-stranded complementary DNA (cDNA) is prepared …
- … copies of which are synthesized using primers and DNA polymerase
Step 6 can be repeated many times, doubling the numbers of DNA molecules created each time; 30 steps, for example, will yield 230 (i.e. 1,073,741,824) or about 109 molecules
An immunoassay has also been described, but it has a high false omission (or exclusion) rate (Table 1).
Table 1. Diagnostic features of an immunoassay for SARS-CoV-2

Efforts are currently being made to develop and implement an immunoassay for antiviral antibodies to determine whether infection has previously occurred.
Appendix 1: Varieties of coronaviruses
Coronaviridae is the name given to a family of viruses with two subfamilies, Letovirinae and Coronavirinae. The latter has four genera, Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus, These include seven coronaviruses that can infect humans (Table 2). Coronaviruses can also infect non-human mammals (Table 3), they can be carried by birds or infect them (Table 4), and they can be carried by bats (Table 5).
Table 2. Taxonomy of coronaviruses that can cause disease in humans
Table 3. Some non-human mammals that can be infected by coronaviruses
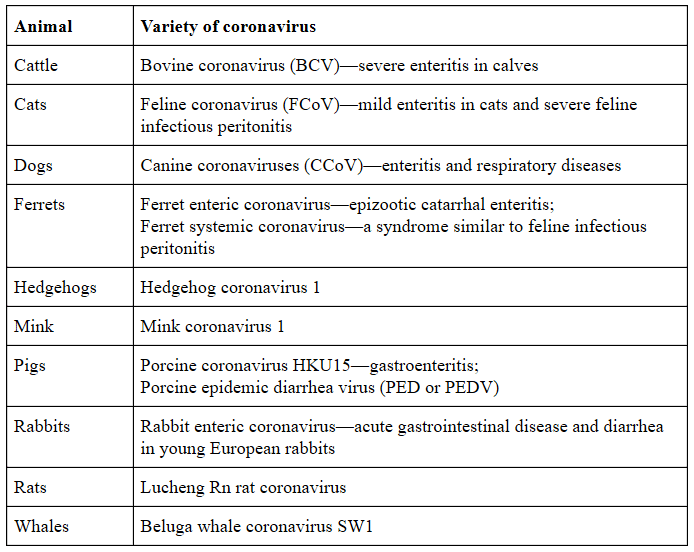
Table 4. Some birds that can carry or be infected by coronaviruses (gammacoronaviruses and deltacoronaviruses)
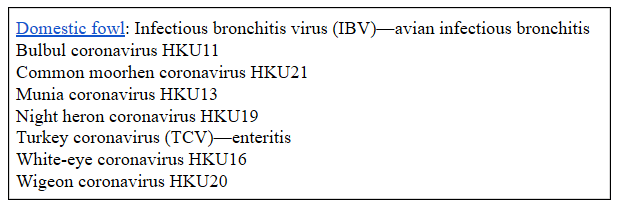
Table 5. Some coronaviruses carried by bats

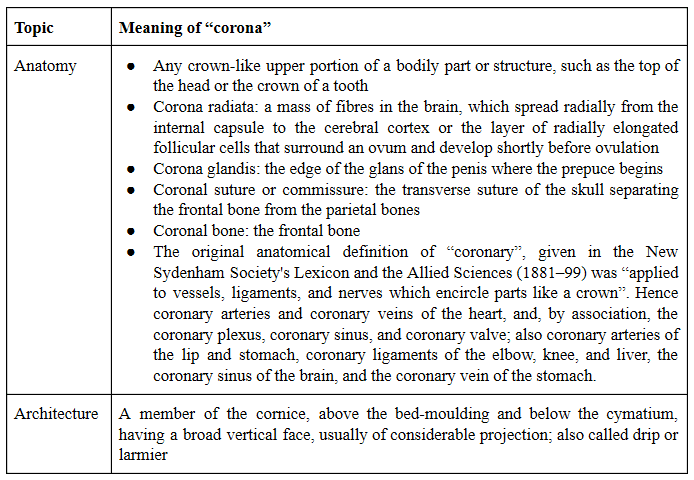
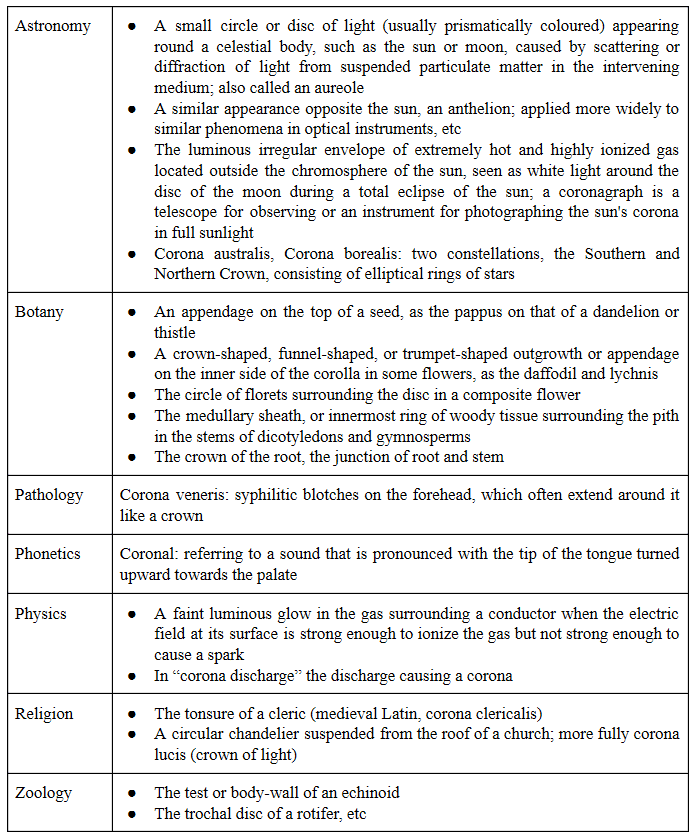
Appendix 2: Meanings of the word “corona”
I have listed the many different meanings of “corona” and some of its derivatives in Table 6 below.
Table 6. Different meanings of “corona” and some derivatives (based on definitions in the Oxford English Dictionary)